Unix shell pipeline: Part 2 - Using temporary file for IPC
This is a continuation of the prior post. I'll describe some of the findings. Recall the sample problem was doing recursive grep in a directory.
Study Solution:
find . ( -path *obj-ppc -o -path *lib-ppc -o -path *bin-ppc -o
-path *\.CC -o -path */\.T* ) -prune -o -type f -print |
xargs file -- | grep text | cut -d: -f1 |
xargs grep $pattern
The solution comprised of using find to generate the whole list of files. Then use the 'file' command to weed out all non-text files. Finally pass the file-list through grep to do the actual search. We also used 'xargs' to ensure minimal copies of processes are forked (for the file and grep).
We already did some optimization. The best ones include using -prune of find to cut branches of uninteresting directories like binaries, object directories. The second was using 'file' command to remove non-text files. When I did the measurement this process was really slow. Sometimes it even took more time than the last grep! I expected file to run pretty fast as I had thought it uses some magic number just in the beginning of the file and does not scan the whole file content.
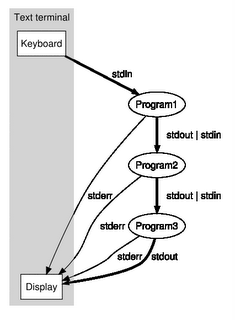
As we saw in the last post, the real killer of speed is the IPC between the processes in the pipeline. I removed the IPC and used temporary files to capture the intermediate results. The performance boost was immense. In fact the same run on 100k files could complete literally under 40 seconds when the IPC based one took forever (I think at least 15 minutes). Moreover the file cache influence was substantial. The first run took about 2 minutes or so; but the immediate next run will complete under 40 seconds. The first run would have a CPU util of about 1% while the second run hitting almost 40% at times. This percentage is the CPU time (both user+sys) versus wall-clock time. I think getting to anything above 25% is impressive.
The next step was I tried to exploit the Dual-CPU and all the 2nd cache resources. Our performance is still a lot I/O bound; but let's try to use all the intermediate caches and RAM. The solution is to take each intermediate temporary file and break it into n pieces. Then feed each of these pieces to n threads of the next stage. Thus we combine multi-threading with temporary file IPCs. The gain was good considering we are still bound by IO. BTW the splitting was done using unix 'split' command (see below in code).
An added bonus of the temporary file approach is re-usability of results from prior run. For our problem, if only the search string changes, say from needle to pin, we can reuse the results of all the prior stages. Thus no need to fire up find or file. Just jump directly to the last stage of the pipeline.
Thus the bottom line observation: File systems with large RAM caches have gone through decades of optimizations - so exploit it. Instead of using IPC through pipes between a producer and consumer, use temporary files. The loss in parallel running of the two processes is well compensated by allowing each to run at its full speed. Each process's output and input are connected to temporary disk files.
Here are some of the code listing.. showing the essential part of using temporary file for IPC and splitting it for parallel processing. Please excuse the formatting.. I had to resort to HTML pre tag.
# command processor using file IO redirection
# A thread which will run a unix command. Objective is to get the
# command complete ASAP. Thus stdin, stdout, stderr are all redirected
# from regular disk files. The guess is with no other process involved,
# the command should be able to run at top speed
#
class command_processor_fileio (Thread):
def __init__ (self, cmd, fname_stdin, fname_out):
Thread.__init__(self)
self.cmd = cmd # Unix cmd.. must be string..sent to Popen
self.fname_stdin = fname_stdin # stdin redirect fname
self.fname_out = fname_out # common prefix to stdout/stderr
self.sh_output = [] # in case intermediate sh complains
self.fname_stdout = '' # set up later in run
self.fname_stderr = '' # set up later in run
self.status = 0 # status of cmd execution
self.status_is_signaled = 0 # status of cmd execution ..is signal?
def run(self):
cmd_run = self.cmd
thread_name = self.getName()
self.fname_stdout = "%s-%s-stdout.txt" % (self.fname_out, thread_name)
self.fname_stderr = "%s-%s-stderr.txt" % (self.fname_out, thread_name)
redirection = ' >|%s 2>|%s < %s' % (self.fname_stdout, self.fname_stderr, self.fname_stdin)
cmd_run += redirection
if options.debug > 0:
print "Starting thread %s cmd %s" % (thread_name, cmd_run)
run = popen2.Popen3(cmd_run)
run.tochild.close() # all redirec. So we don't expect any input
# following may be skipped if it aids in performance
self.sh_output = run.fromchild.readlines() # just in case sh complains
result = run.wait() # block wait until cmd is finished.
self.status = os.WEXITSTATUS(result)
self.status_is_signaled = os.WIFSIGNALED(result)
#
# Merge all the output of various threads into one file
#
def thread_output_files_merge (stdout_files, stderr_files, target):
fname_stdout = redirection_filename(target)
fname_stderr = redirection_filename(target, is_stderr=True)
# just merge all files.. so do a cat f1 f2 f3 ...fn > f_out
merge_cmd = "cat %s > %s" % ( ' '.join(stdout_files), fname_stdout)
os.system(merge_cmd)
merge_cmd = "cat %s > %s" % ( ' '.join(stderr_files), fname_stderr)
os.system(merge_cmd)
#
# Split one file into many smaller files for each thread to work on
# The routine must return num_threads files... even empty ones are okay
#
def thread_input_file_split (fname, num_threads):
if num_threads == 1:
# easiest to just get out early
return [fname]
fsize = os.path.getsize(fname)
bytes_per_file = fsize / num_threads
split_prefix = "%s-thread-" % (fname,)
if options.use_shortcut > 1: # User gave -s -s .. that means don't
# do the split of files.. just re-use from prior run even the split
# files. NOTE: -t value must remain the same as in prior run
split_files = glob.glob(split_prefix + 'a?') # split.. uses aa..ab.ac
# must have atleast num_threads elements.
if options.debug > 0:
print "Reusing thread split files (%d files) from prior run " "due to -s -s: %s" % (num_threads, split_files)
return split_files[:num_threads]
# before running the split command, remove any old split output files
# from a prior run
old_files = glob.glob(split_prefix + 'a?')# man split.. uses aa..ab
for old_file in old_files:
if options.debug > 0:
print "Previous run leftovers. Removing ..", old_file
os.remove(old_file)
split_cmd = "split -C%d %s %s" % (bytes_per_file, fname, split_prefix)
os.system(split_cmd)
# Assert: We will have at least num_threads now .. not less
split_files = glob.glob(split_prefix + 'a?') # man split.. uses aa..ab.ac
split_files.sort() # exploit aa..ab..naming.. ais the smallest file
while len(split_files) > num_threads:
# Take the last file (smallest one) and merge it with it's prior file
smallest_file = split_files.pop()
merge_cmd = "cat %s >> %s" % (smallest_file, split_files[-1])
if options.debug > 0:
print "Merging smallest file: cmd %s" % (merge_cmd, )
os.system(merge_cmd)
if options.debug > 0:
print "Multiple thread input split_files: ", split_files
# ASSERT: len(split_files) == num_threads
return split_files
#
# Run cmd using given number of threads
#
def execute_child_threads_fileio (cmd, stdin_fname, num_threads, target):
stdin = file_get_all_lines(stdin_fname)
stdin_size = len(stdin)
min_per_thread = 4096 # just some minimum size for a thread
fsize = os.path.getsize(stdin_fname)
if fsize < min_per_thread: # must be * num_threads.. but okay
num_threads = 1
#split the file into num_thread pieces
stdin_fnames = thread_input_file_split(stdin_fname, num_threads)
fname_out = redirection_filename(target, is_prefix=True)
#work_slice = stdin_size / num_threads; # each thread's work size
workers = []
# We are going to chop stdin into equal sized chunks for each thread.
for i in range(num_threads):
#start_index = i * work_slice;
#end_index = start_index + work_slice;
#if i == num_threads - 1: # if last worker...take all remaining
# end_index = stdin_size;
#this_stdin = stdin[start_index:end_index] # this thread's in data
cp = command_processor_fileio(cmd, stdin_fnames[i], fname_out)
workers.append(cp);
cp.start() # fire up the thread to do the data munching
stdout_files = [] # collect each thread's output file
stderr_files = []
for worker in workers:
worker.join() # wait for each worker to finish
#TODO. Handle each thread's return status...
stdout_files.append(worker.fname_stdout)
stderr_files.append(worker.fname_stderr)
# merge all its output files into one.. so that next stage process
# in the pipeline sees one file.
thread_output_files_merge(stdout_files, stderr_files, target)
return ([] , 0, 0)